Hi Probeman,
I followed all your recommendations but not understood, how I can work with more then 1 point in CalcZaf.
I want to use 2 options:
1. use the CalcZAF export menu from Probe for EPMA - I saw just 1 point in CalcZaf table, how I can see more then one?
2. manually input to CalcZaf my data (from JEOL soft? for instance) - everything is clear if I calculated one point, but how I can calculate more then one.
Hi Rom,
I had to re-read this topic to see what you exactly are trying to accomplish and it seems you are wanting to re-calculate all analysis points in Probe for EPMA using all 10 matrix corrections. If that is what you want to do, why don't you just use the Use All Matrix Corrections" checkbox as described here:
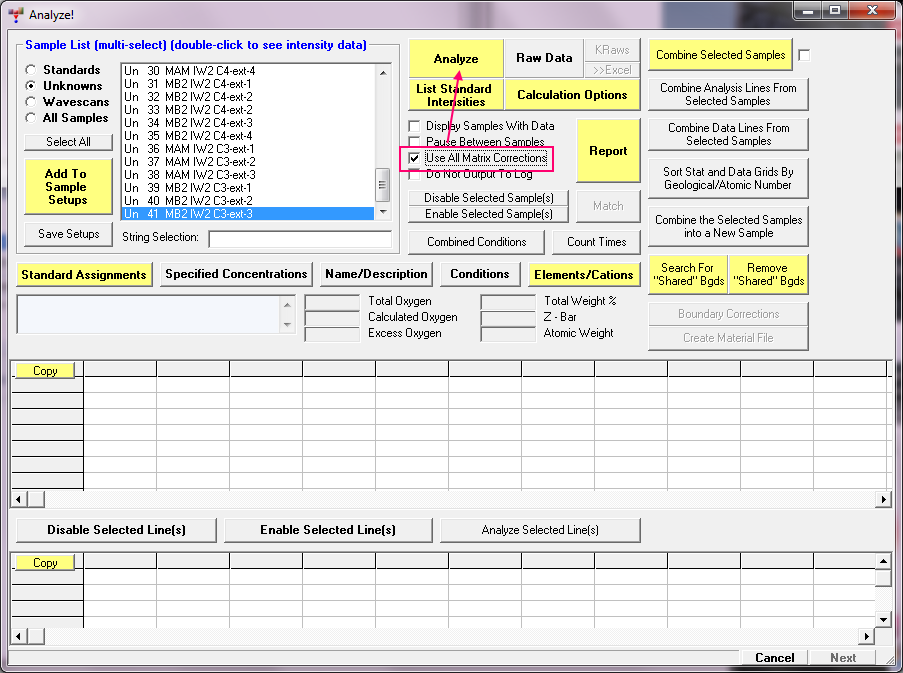
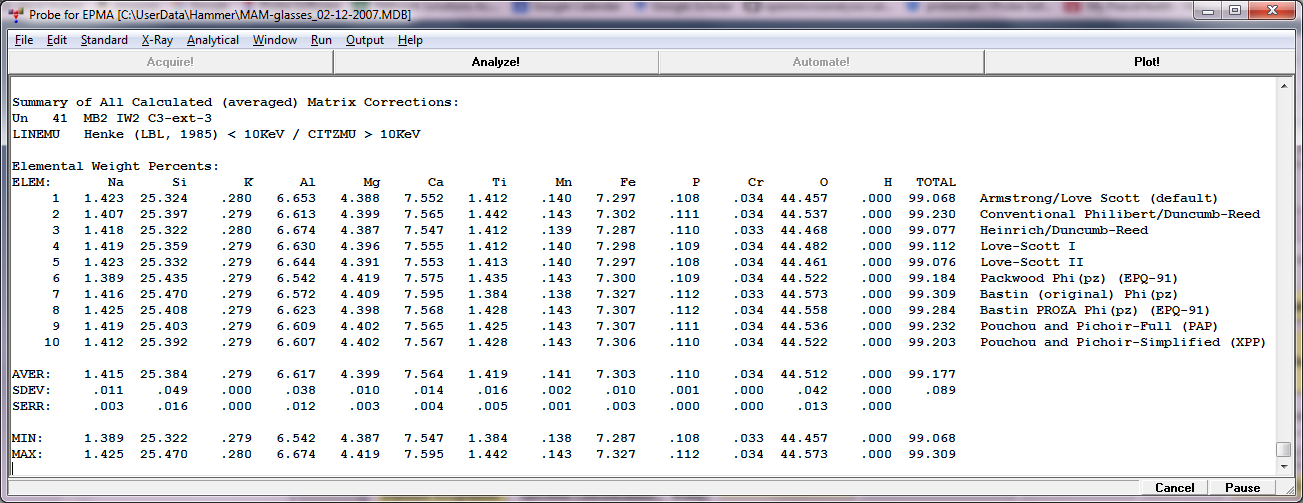
and then click the Analyze! button and you'll get the output for the 10 matrix corrections in the log window:
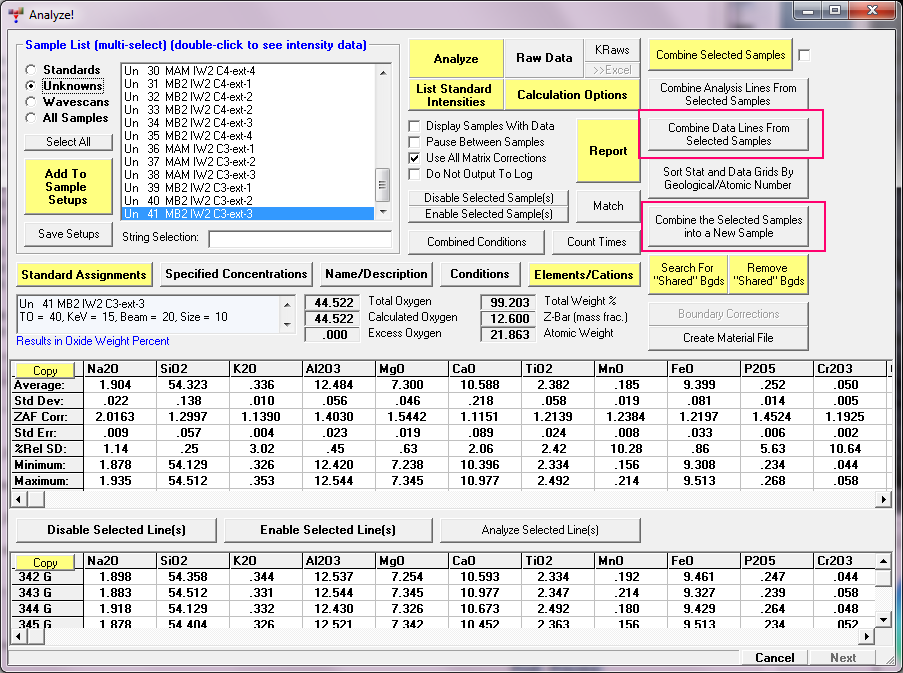
Remember you can also combine points from multiple samples in PFE (up to 500 points) using these buttons:
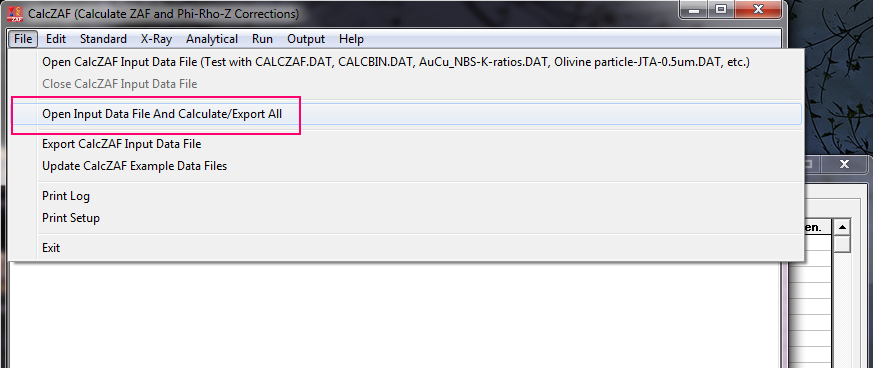
As for CalcZAF, if you really want to re-process your PFE data in CalcZAF (why?), you can certainly export all the data using a right click in the Analyze! window or from the Output menu, but after importing your data into CalcZAF, it only displays one point at a time. Then use the Load Next Dataset From Input File, to see the next point.
To have CalcZAF re-calculate all the points in the imported DAT file, simply use this menu to open the .DAT file:
But I think you're better off just using Probe for EPMA.