One of the cool features in Probe for EPMA is that the program can properly handle the presence of "duplicate" elements for fully quantitative analysis.
This feature is often utilized with the "aggregate" mode, for increasing the "geometric efficiency" of the instrument by combining photons (in software!) from different spectrometers to improve sensitivity. Note however, for this "aggregate" mode, the software will only combine photons from elements that utilize the *same* x-ray emission line! Why? Because the physics doesn't care about the analyzing crystals used (photons is photons!), but the photon energies are very important for the physics, so they cannot be combined at the photon level.
Of course one might also just want some replicate measurements on multiple spectrometers without combining them using "aggregate" mode. If these are trace elements they will not affect the matrix correction enough to matter so that is fine. But if the concentrations of the duplicate elements are large enough, the matrix correction will be adversely affected.
Let's look at an example of this, using some GaAs and GaSb synthetic standards. Here is the output from the analysis of GaSb:
St 669 Set 2 GaSb (synthetic)
TakeOff = 40.0 KiloVolt = 20.0 Beam Current = 20.0 Beam Size = 10
(Magnification (analytical) = 40000), Beam Mode = Analog Spot
(Magnification (default) = 400, Magnification (imaging) = 800)
Image Shift (X,Y): .00, .00
(MAC block)
Number of Data Lines: 5 Number of 'Good' Data Lines: 5
First/Last Date-Time: 01/07/2014 05:39:27 PM to 01/07/2014 05:47:40 PM
WARNING- Using Exponential Off-Peak correction for as ka
WARNING- Using Exponential Off-Peak correction for sb la
Average Total Oxygen: .000 Average Total Weight%: 154.313
Average Calculated Oxygen: .000 Average Atomic Number: 39.191
Average Excess Oxygen: .000 Average Atomic Weight: 84.510
Average ZAF Iteration: 5.00 Average Quant Iterate: 2.00
WARNING- Duplicate analyzed elements are present in the sample matrix!!
Use Aggregate Intensity option or Disable Quant feature for accurate matrix correction.
St 669 Set 2 GaSb (synthetic), Results in Elemental Weight Percents
ELEM: Ga Ga Ga As In Sb
BGDS: LIN LIN LIN EXP LIN EXP
TIME: 60.00 60.00 60.00 60.00 30.00 30.00
BEAM: 19.94 19.94 19.94 19.94 19.94 19.94
ELEM: Ga Ga Ga As In Sb SUM
XRAY: (la) (ka) (la) (ka) (la) (la)
137 28.309 34.369 28.601 -.038 -.059 63.473 154.656
138 28.262 34.325 28.414 -.144 -.040 63.320 154.137
139 28.303 34.444 28.558 -.097 -.070 63.177 154.315
140 28.363 34.395 28.473 -.038 -.096 63.243 154.339
141 28.368 34.292 28.484 -.080 -.051 63.107 154.120
AVER: 28.321 34.365 28.506 -.079 -.063 63.264 154.313
SDEV: .044 .059 .074 .044 .022 .141 .216
SERR: .020 .026 .033 .020 .010 .063
%RSD: .16 .17 .26 -55.91 -34.07 .22
PUBL: 36.413 36.413 36.413 n.a. n.a. 63.587 100.000
%VAR: -22.22 -5.62 -21.72 --- --- (-.51)
DIFF: -8.092 -2.048 -7.907 --- --- (-.32)
STDS: 662 662 662 662 2002 669
STKF: .4679 .5103 .4679 .4873 .5621 .5907
STCT: 648.92 694.60 232.91 91.23 200.87 236.11
UNKF: .1403 .3532 .1413 -.0008 -.0006 .5906
UNCT: 194.62 480.73 70.31 -.15 -.21 236.09
UNBG: 2.62 12.42 .73 3.04 1.66 2.79
ZCOR: 2.0181 .9730 2.0181 1.0192 1.0841 1.0711
KRAW: .2999 .6921 .3019 -.0016 -.0010 1.0000
PKBG: 75.25 39.72 97.62 .95 .88 85.59
INT%: ---- ---- ---- ---- ---- ----
First note that we are acquiring Ga La on two spectrometers and Ga Ka on one spectrometer, plus As, In and Sb. GaSb is the primary standard for Sb and GaAs is the primary standard for Ga.
Next we note that the analysis is crazy. The total is over 150% and we have three times as much Ga as we should... we makes sense because we are measuring Ga on three spectrometers! So let's begin by turning on the "aggregate" mode in the Analytical | Analysis Options dialog as seen here:
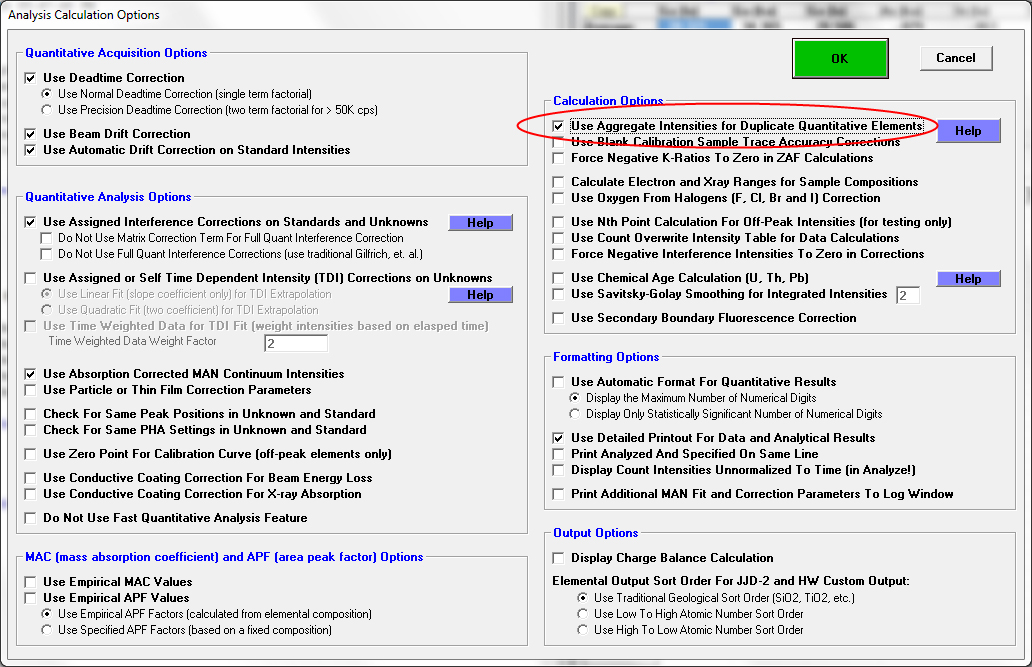
Now we get this output when we click the Analyze button:
St 669 Set 2 GaSb (synthetic)
TakeOff = 40.0 KiloVolt = 20.0 Beam Current = 20.0 Beam Size = 10
(Magnification (analytical) = 40000), Beam Mode = Analog Spot
(Magnification (default) = 400, Magnification (imaging) = 800)
Image Shift (X,Y): .00, .00
(MAC block)
Number of Data Lines: 5 Number of 'Good' Data Lines: 5
First/Last Date-Time: 01/07/2014 05:39:27 PM to 01/07/2014 05:47:40 PM
WARNING- Using Exponential Off-Peak correction for as ka
WARNING- Using Exponential Off-Peak correction for sb la
WARNING- Using Aggregate Intensities for Duplicate Elements
Element ga la on channel 1(LTAP) is duplicated by ga la on channel 3(TAP)
Element ga la on channel 1(LTAP) is duplicated by ga la on channel 3(TAP)
Average Total Oxygen: .000 Average Total Weight%: 129.446
Average Calculated Oxygen: .000 Average Atomic Number: 40.787
Average Excess Oxygen: .000 Average Atomic Weight: 88.152
Average ZAF Iteration: 5.00 Average Quant Iterate: 2.00
St 669 Set 2 GaSb (synthetic), Results in Elemental Weight Percents
ELEM: Ga Ga Ga As In Sb
BGDS: LIN LIN LIN EXP LIN EXP
TIME: 60.00 60.00 .00 60.00 30.00 30.00
BEAM: 19.94 19.94 .00 19.94 19.94 19.94
AGGR: 2
ELEM: Ga Ga Ga As In Sb SUM
XRAY: (la) (ka) (la) (ka) (la) (la)
137 31.221 34.985 .000 -.038 -.059 63.615 129.725
138 31.123 34.941 .000 -.143 -.039 63.462 129.343
139 31.202 35.062 .000 -.097 -.070 63.319 129.417
140 31.220 35.011 .000 -.038 -.095 63.387 129.485
141 31.233 34.907 .000 -.080 -.050 63.249 129.259
AVER: 31.200 34.981 .000 -.079 -.063 63.406 129.446
SDEV: .044 .060 .000 .044 .021 .141 .177
SERR: .020 .027 .000 .020 .010 .063
%RSD: .14 .17 .09 -55.91 -34.07 .22
PUBL: 36.413 36.413 n.a. n.a. n.a. 63.587 100.000
%VAR: -14.32 -3.93 .00 --- --- (-.28)
DIFF: -5.213 -1.432 .00 --- --- (-.18)
STDS: 662 662 0 662 2002 669
STKF: .4637 .5222 .0000 .4908 .5621 .5976
STCT: 881.83 694.60 .00 91.23 200.87 236.11
UNKF: .1393 .3614 .1413 -.0008 -.0006 .5976
UNCT: 264.92 480.73 .00 -.15 -.21 236.09
UNBG: 3.35 12.42 .00 3.04 1.66 2.79
ZCOR: 2.2399 .9679 2.0181 1.0065 1.0729 1.0610
KRAW: .3004 .6921 .3019 -.0016 -.0010 1.0000
PKBG: 80.05 39.72 .00 .95 .88 85.59
INT%: ---- ---- ---- ---- ---- ----
This is a little better because the two Ga La channels have been "aggregated" as we specified, but we still have duplicate Ga La and Ga Ka. What we need to do here is to "turn off" either the 2 Ga La channels or the single Ga Ka channel using the "Disable Quant" feature.
Since the Ga La channels are being "aggregated" it is critically important to use the "Disable Quant" feature on *both* the standards *and* the unknowns. Since this is a secondary standard for Ga, we would want to apply the "Disable Quant" flags to both the primary GaAs standard and the GaSb secondary standard.
Or we could just set the "Disable Quant" flag for *only* the Ga Ka channel in this standard sample as seen here:
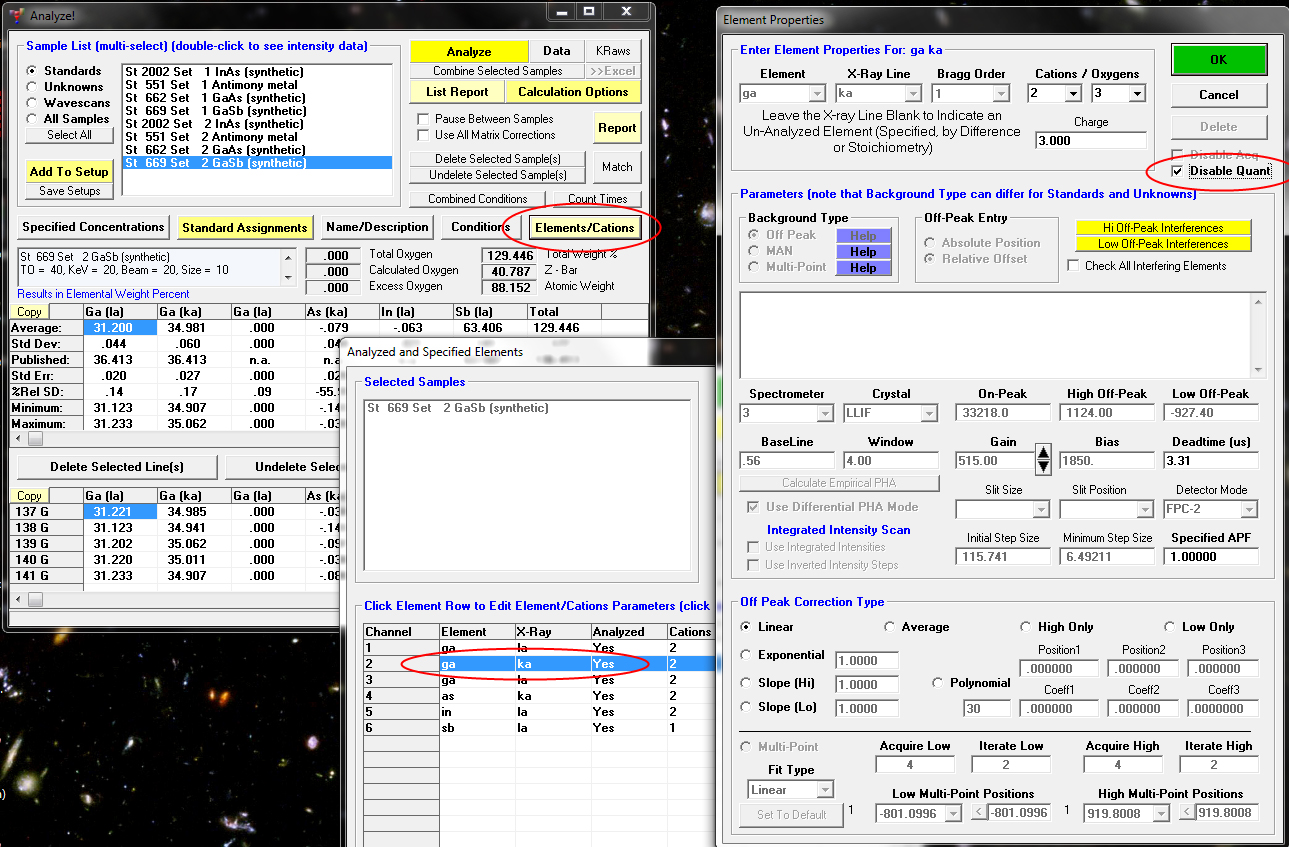
since it is not duplicated as far as the "aggregate" mode is concerned.
Now, when we again click the Analyze button in the Analyze! window we obtain these reults:
St 669 Set 2 GaSb (synthetic)
TakeOff = 40.0 KiloVolt = 20.0 Beam Current = 20.0 Beam Size = 10
(Magnification (analytical) = 40000), Beam Mode = Analog Spot
(Magnification (default) = 400, Magnification (imaging) = 800)
Image Shift (X,Y): .00, .00
(MAC block)
Number of Data Lines: 5 Number of 'Good' Data Lines: 5
First/Last Date-Time: 01/07/2014 05:39:27 PM to 01/07/2014 05:47:40 PM
WARNING- Using Exponential Off-Peak correction for as ka
WARNING- Using Exponential Off-Peak correction for sb la
WARNING- Using Aggregate Intensities for Duplicate Elements
Element ga la on channel 1(LTAP) is duplicated by ga la on channel 3(TAP)
Element ga la on channel 1(LTAP) is duplicated by ga la on channel 3(TAP)
WARNING- Quantitation is Disabled For ga ka, Spectro 3
Average Total Oxygen: .000 Average Total Weight%: 99.891
Average Calculated Oxygen: .000 Average Atomic Number: 43.717
Average Excess Oxygen: .000 Average Atomic Weight: 95.732
Average ZAF Iteration: 7.00 Average Quant Iterate: 2.00
St 669 Set 2 GaSb (synthetic), Results in Elemental Weight Percents
ELEM: Ga Ga Ga As In Sb
BGDS: LIN LIN LIN EXP LIN EXP
TIME: 60.00 --- .00 60.00 30.00 30.00
BEAM: 19.94 --- .00 19.94 19.94 19.94
AGGR: 2 ---
ELEM: Ga Ga-D Ga As In Sb SUM
XRAY: (la) (ka) (la) (ka) (la) (la)
137 36.465 --- .000 -.038 -.057 63.792 100.162
138 36.363 --- .000 -.142 -.039 63.634 99.817
139 36.467 --- .000 -.096 -.068 63.491 99.794
140 36.475 --- .000 -.037 -.094 63.562 99.905
141 36.479 --- .000 -.079 -.049 63.425 99.776
AVER: 36.450 --- .000 -.078 -.061 63.581 99.891
SDEV: .049 --- .000 .044 .021 .141 .159
SERR: .022 --- .000 .020 .009 .063
%RSD: .13 --- .09 -55.91 -34.07 .22
PUBL: 36.413 n.a. n.a. n.a. n.a. 63.587 100.000
%VAR: .10 .00 .00 --- --- (-.01)
DIFF: .037 .00 .00 --- --- (-.01)
STDS: 662 --- 0 662 2002 669
STKF: .4557 --- .0000 .4976 .5621 .6098
STCT: 881.83 --- .00 91.23 200.87 236.11
UNKF: .1369 --- .0000 -.0008 -.0006 .6097
UNCT: 264.92 --- .00 -.15 -.21 236.09
UNBG: 3.35 --- .00 3.04 1.66 2.79
ZCOR: 2.6622 --- .0000 .9833 1.0529 1.0428
KRAW: .3004 --- .0000 -.0016 -.0010 1.0000
PKBG: 80.05 --- .00 .95 .88 85.59
INT%: ---- --- ---- ---- ---- ----
See the line above labeled VAR% which is the relative percent variance from the "published" value. Of course a relative accuracy of less than 1% is almost too good to be true, but we'll take it! :-)
Especially since the matrix correction for Ga La in this standard is over 200%, so perhaps we ought to call it "spurious accuracy".